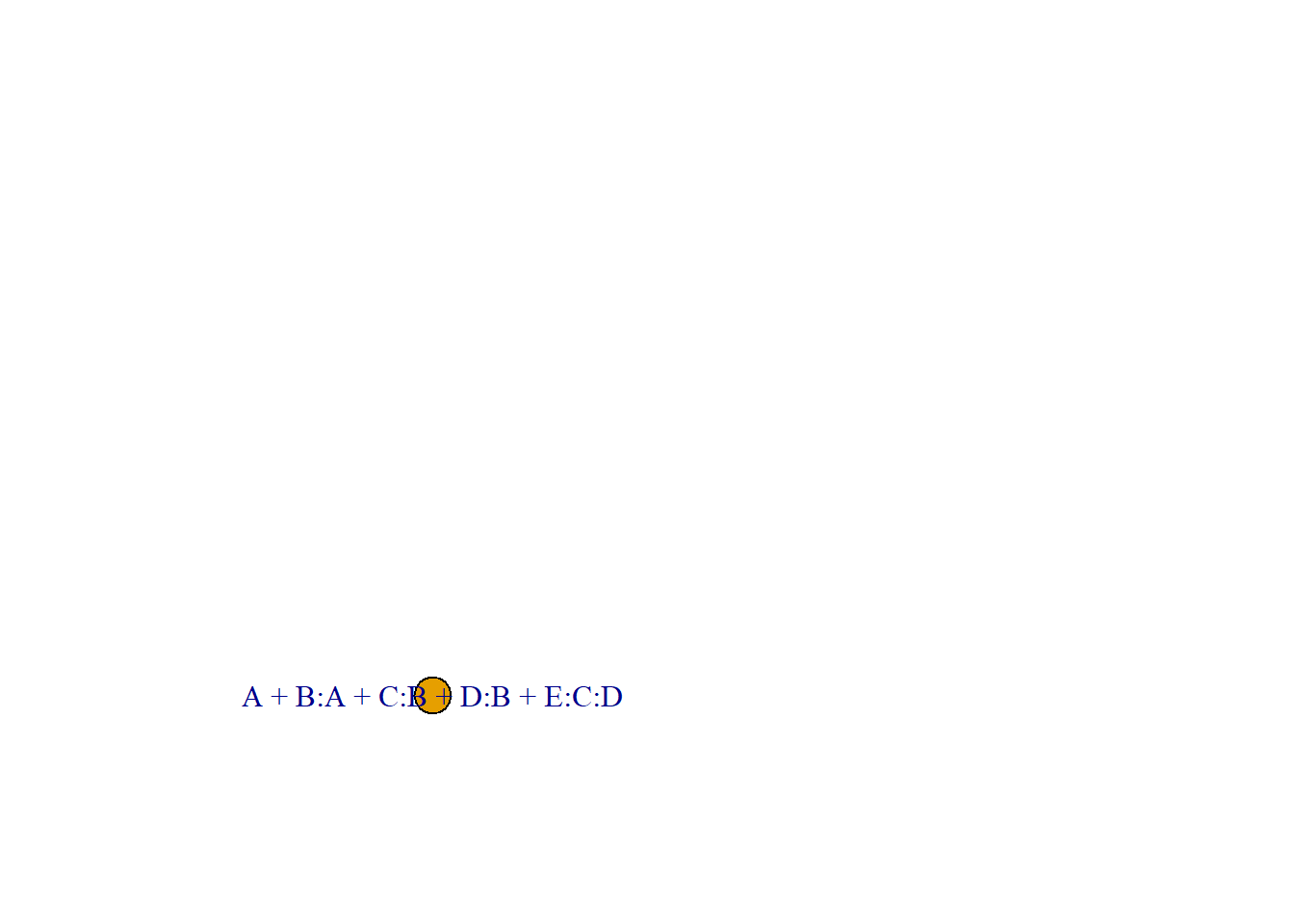install.packages("gRain")贝叶斯与概率图模型(PGM)
1 概率图模型(Probabilistic Graphical Models)
概率图模型是一种结合概率论与图论的理论框架,用于表示和分析复杂系统中的不确定性及概率关系。
1.1 常见类型
贝叶斯网络
马尔科夫随机场(MRF)
隐马尔科夫模型(HMM)
1.2 用R来展示概率图模型
1.2.1 使用R程序包gRain。
安装:
加载:
首先定义一个带有变量的A、B、C、D、E的简单无向图
安装可视化程序包,使用流行的Rgraphviz
Rgraphviz是 Bioconductor 生态系统的一部分,所以可以通过 Bioconductor 来安装它。首先要确保已经安装了BiocManager,若未安装,可使用以下代码进行安装:
if (!require("BiocManager", quietly = TRUE))
install.packages("BiocManager")安装好BiocManager之后,使用它来安装Rgraphviz:
BiocManager::install("Rgraphviz")载入并使用:
定义有向图:
1.3 报错
报错:无法正确显示图形
原因:Rgraphviz 没有被正确的安装。
需要从 bioconductor 中安装:
> install.packages("Rgraphviz")
Installing package into 'C:/Users/asus/AppData/Local/R/win-library/4.4'
(as 'lib' is unspecified)
Warning message:
package 'Rgraphviz' is not available for this version of R
A version of this package for your version of R might be available elsewhere,
see the ideas at
https://cran.r-project.org/doc/manuals/r-patched/R-admin.html#Installing-packages当使用 bidconductor 源来安装 Rgraphviz ,首先安装 BiocManager :
> if (!require("BiocManager", quietly = TRUE))
+ install.packages("BiocManager")
Installing package into 'C:/Users/asus/AppData/Local/R/win-library/4.4'
(as 'lib' is unspecified)
trying URL 'https://mirrors.sustech.edu.cn/CRAN/bin/windows/contrib/4.4/BiocManager_1.30.25.zip'
Content type 'application/zip' length 506482 bytes (494 KB)
downloaded 494 KB
package 'BiocManager' successfully unpacked and MD5 sums checked
The downloaded binary packages are in
C:\Users\asus\AppData\Local\Temp\RtmpSCcZNG\downloaded_packages然后使用 BiocManager 安装 Rgraphviz ,需要安装的配套程序包很多,需要几分钟,但是这里出现了报错。
> BiocManager::install("Rgraphviz")
'getOption("repos")' replaces Bioconductor standard repositories, see
'help("repositories", package = "BiocManager")' for details.
Replacement repositories:
CRAN: https://mirrors.sustech.edu.cn/CRAN
Bioconductor version 3.20 (BiocManager 1.30.25), R 4.4.2 (2024-10-31 ucrt)
Installing package(s) 'BiocVersion', 'Rgraphviz'
also installing the dependencies 'BiocGenerics', 'graph'
trying URL 'https://bioconductor.org/packages/3.20/bioc/bin/windows/contrib/4.4/BiocGenerics_0.52.0.zip'
libpng warning: iCCP: known incorrect sRGB profile
libpng warning: iCCP: known incorrect sRGB profile
Content type 'application/zip' length 639558 bytes (624 KB)
downloaded 624 KB
trying URL 'https://bioconductor.org/packages/3.20/bioc/bin/windows/contrib/4.4/graph_1.84.1.zip'
Content type 'application/zip' length 2173761 bytes (2.1 MB)
downloaded 2.1 MB
trying URL 'https://bioconductor.org/packages/3.20/bioc/bin/windows/contrib/4.4/BiocVersion_3.20.0.zip'
Content type 'application/zip' length 8386 bytes
downloaded 8386 bytes
trying URL 'https://bioconductor.org/packages/3.20/bioc/bin/windows/contrib/4.4/Rgraphviz_2.50.0.zip'
Content type 'application/zip' length 1457153 bytes (1.4 MB)
downloaded 1.4 MB
package 'BiocGenerics' successfully unpacked and MD5 sums checked
package 'graph' successfully unpacked and MD5 sums checked
package 'BiocVersion' successfully unpacked and MD5 sums checked
package 'Rgraphviz' successfully unpacked and MD5 sums checked
The downloaded binary packages are in
C:\Users\asus\AppData\Local\Temp\RtmpSCcZNG\downloaded_packages
Installation paths not writeable, unable to update packages
path: C:/Program Files/R/R-4.4.2/library
packages:
class, cli, cluster, curl, data.table, foreign, glue, jsonlite, KernSmooth,
lintr, MASS, Matrix, nlme, nnet, processx, ps, purrr, R.utils, R6, renv,
reticulate, rlang, rpart, sessioninfo, spatial, survival, tinytex, xfun,
xml2, zoo
Old packages: 'httpgd', 'readxl', 'unigd'
apdate all/some/none? [a/s/n]:
trying URL 'https://mirrors.sustech.edu.cn/CRAN/bin/windows/contrib/4.4/httpgd_2.0.3.zip'
Content type 'application/zip' length 874316 bytes (853 KB)
downloaded 853 KB
trying URL 'https://mirrors.sustech.edu.cn/CRAN/bin/windows/contrib/4.4/readxl_1.4.4.zip'
Content type 'application/zip' length 750422 bytes (732 KB)
downloaded 732 KB
trying URL 'https://mirrors.sustech.edu.cn/CRAN/bin/windows/contrib/4.4/unigd_0.1.3.zip'
Content type 'application/zip' length 5591347 bytes (5.3 MB)
downloaded 5.3 MB
package 'httpgd' successfully unpacked and MD5 sums checked
package 'readxl' successfully unpacked and MD5 sums checked
Warning: cannot remove prior installation of package 'readxl'
Warning: restored 'readxl'
package 'unigd' successfully unpacked and MD5 sums checked
The downloaded binary packages are in
C:\Users\asus\AppData\Local\Temp\RtmpSCcZNG\downloaded_packages
Warning message:
In file.copy(savedcopy, lib, recursive = TRUE) :
problem copying C:\Users\asus\AppData\Local\R\win-library\4.4\00LOCK\readxl\libs\x64\readxl.dll to C:\Users\asus\AppData\Local\R\win-library\4.4\readxl\libs\x64\readxl.dll: Permission denied主要问题是没有写入权限,这可能和在 vscode 中安装有关,因此,换到 R 的终端中进行安装(注意,可能需要使用管理员权限),但是仍然免不了一系列的包冲突问题,需要根据提示选择操作:
使用管理员身份运行 R 。
选择合适的源来下载。
-
处理有冲突的包(卸载),或者重新创建一个 env 。
# 删除 readxl 包 readxl_path <- system.file(package = "readxl") if (!is.na(readxl_path)) { unlink(readxl_path, recursive = TRUE) } # 删除 cli 包 cli_path <- system.file(package = "cli") if (!is.na(cli_path)) { unlink(cli_path, recursive = TRUE) } # 再次尝试安装 Rgraphviz BiocManager::install("Rgraphviz") 强制更新,忽略冲突
有些包的版本已经是最新的,但安装过程仍然提示需要更新,你可以使用 force = TRUE 参数强制重新安装。
BiocManager::install("Rgraphviz", force = TRUE)-
具体信息如下:
> BiocManager::install("Rgraphviz") 'getOption("repos")' replaces Bioconductor standard repositories, see 'help("repositories", package = "BiocManager")' for details. Replacement repositories: CRAN: https://mirrors.nju.edu.cn/CRAN Bioconductor version 3.20 (BiocManager 1.30.25), R 4.4.2 (2024-10-31 ucrt) Warning message: package(s) not installed when version(s) same as or greater than current; use `force = TRUE` to re-install: 'Rgraphviz' > BiocManager::install("Rgraphviz", force = TRUE) 'getOption("repos")' replaces Bioconductor standard repositories, see 'help("repositories", package = "BiocManager")' for details. Replacement repositories: CRAN: https://mirrors.nju.edu.cn/CRAN Bioconductor version 3.20 (BiocManager 1.30.25), R 4.4.2 (2024-10-31 ucrt) Installing package(s) 'Rgraphviz' trying URL 'https://bioconductor.org/packages/3.20/bioc/bin/windows/contrib/4.4/Rgraphviz_2.50.0.zip' Content type 'application/zip' length 1457153 bytes (1.4 MB) downloaded 1.4 MB package ‘Rgraphviz’ successfully unpacked and MD5 sums checked The downloaded binary packages are in C:\Users\asus\AppData\Local\Temp\Rtmp2j7SlA\downloaded_packages
1.4 灯泡机的简单概率图模型
首先,为每一个节点定义取值:
为两个随机变量定义百分比数值:
使用 gRain 定义随机变量:
这里的 cptable 表示条件概率表1:它是离散型随机变量概率分布的内存表示2。
1.4.1 构建新的概率图模型
plist <- compileCPT(list(M,L))
plist输出结果如上,这里可以清楚地看到之前定义的概率分布
2025.04.05 再次尝试复现程序,失败,暂时停止概率图R程序的复现工作。
end.